Mean Value T Test¶
STMV Class¶
Calculate the mean T-test for a single sample and the mean T-test for two samples
Parameter:¶
data1 (np.ndarray or xr.DataArray): test data1
data2 (np.ndarray or xr.DataArray): test data2, if data2 is None, function will use one sample test.
wrap (bool): if data are wrapped to original xarray coords. Defaults to True.
*param: parameter transports to sts.ttest_1samp or sts.ttest_2samp
**kwargs: parameter transports to sts.ttest_1samp or sts.ttest_2samp
[1]:
import sacpy as scp
import numpy as np
import matplotlib.pyplot as plt
sst = scp.load_sst()["sst"]
ssta = scp.get_anom(sst, method=0)
ssta_djf = scp.XrTools.spec_moth_yrmean(ssta,[12,1,2])
Nino34 = ssta_djf.loc[:, -5:5, 190:240].mean(axis=(1, 2))
[13]:
select1 = (Nino34 >= 0.8) & (Nino34 < 1.1)
ssta_sl1 = ssta_djf[select1]
select2 = Nino34 >= 1.3
ssta_sl2 = ssta_djf[select2]
# 2 examples ttest
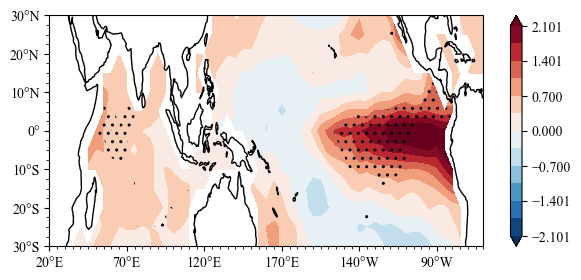
ttest = scp.STMV(ssta_sl2,ssta_sl1)
[14]:
import sacpy.Map
plt.figure(figsize=[7, 3])
m,ax = ttest.mean.splot()
ax.sig_plot(sst.lon,sst.lat,ttest.p_value)
[14]:
<cartopy.mpl.contour.GeoContourSet at 0x15da956d0>
one_mean_test¶
Parameter:¶
data (np.ndarray): test data
expected_mean (np.ndarray, optional): expected mean. Defaults to None.
*param: parameter transports to sts.ttest_1samp
Raises:¶
ValueError: expected_mean shape can match data
Returns:¶
origin_mean,pvalue : np.ndarray: origin_mean and res.value
Example¶
[8]:
import sacpy as scp
import numpy as np
import matplotlib.pyplot as plt
sst = scp.load_sst()["sst"]
ssta = scp.get_anom(sst, method=0)
ssta_djf = scp.XrTools.spec_moth_yrmean(ssta,[12,1,2])
[9]:
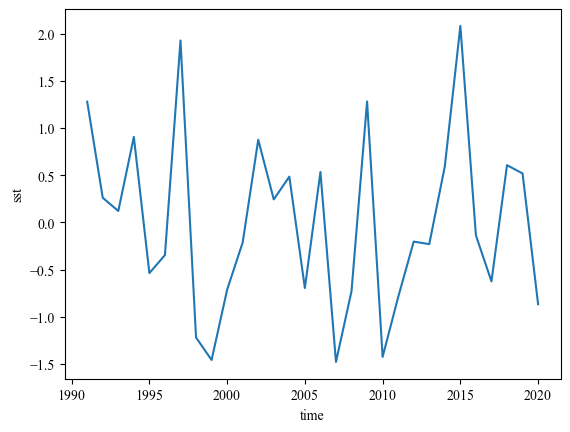
Nino34 = ssta_djf.loc[:, -5:5, 190:240].mean(axis=(1, 2))
print(Nino34.std())
Nino34.plot()
<xarray.DataArray 'sst' ()>
array(0.93506366)
[9]:
[<matplotlib.lines.Line2D at 0x15c593550>]
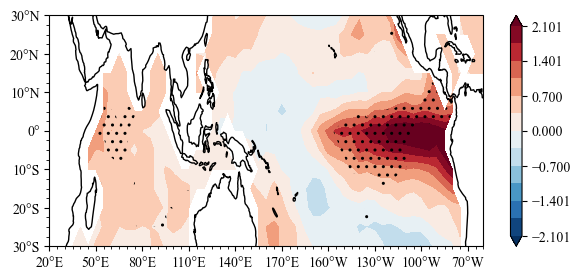
[10]:
select = Nino34 >= 1
ssta_sl = ssta_djf[select]
mean, pv = scp.one_mean_test(np.array(ssta_sl))
[11]:
import sacpy.Map
import cartopy.crs as ccrs
fig = plt.figure(figsize=[7, 3])
ax = plt.axes(projection=ccrs.PlateCarree(central_longitude=180))
lon ,lat = np.array(ssta.lon) , np.array(ssta.lat)
m = ax.scontourf(lon,lat,mean)
n = ax.sig_plot(lon,lat,pv,color="k",marker="..")
ax.init_map(stepx=50, smally=2.5)
plt.colorbar(m)
[11]:
<matplotlib.colorbar.Colorbar at 0x15c648b20>
two_mean_test¶
two samples t test
Parameter¶
data1 (np.ndarray): test data1
data2 (np.ndarray): test data2
Returns:¶
mean_diff (np.ndarray): mean difference
pvalue (np.ndarray): p-value of t-test
Example¶
Load Modules and SST data¶
[16]:
import sacpy as scp
import numpy as np
import matplotlib.pyplot as plt
sst = scp.load_sst()["sst"]
ssta = scp.get_anom(sst, method=0)
ssta_djf = scp.XrTools.spec_moth_yrmean(ssta,[12,1,2])
Nino34 = ssta_djf.loc[:, -5:5, 190:240].mean(axis=(1, 2))
Select Normal and Super El nino Cases¶
[14]:
select1 = (Nino34 >= 0.8) & (Nino34 < 1.1)
ssta_sl1 = ssta_djf[select1]
select2 = Nino34 >= 1.3
ssta_sl2 = ssta_djf[select2]
# cal difference
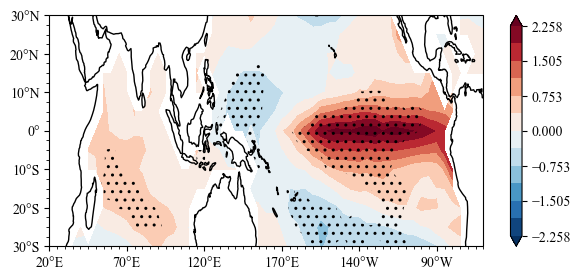
mean_diff, pv2 = scp.two_mean_test(ssta_sl2,ssta_sl1)
Plot the results¶
[15]:
fig = plt.figure(figsize=[7, 3])
ax = plt.axes(projection=ccrs.PlateCarree(central_longitude=180))
lon ,lat = np.array(ssta.lon) , np.array(ssta.lat)
m = ax.scontourf(lon,lat,mean_diff)
n = ax.sig_plot(lon,lat,pv2,color="k",marker="..")
ax.init_map(stepx=50, smally=2.5)
plt.colorbar(m)
[15]:
<matplotlib.colorbar.Colorbar at 0x15cae6730>